Metran practical example
This notebook shows a practical application of Metran on calculated residuals from univariate time series models as published in the article van Geer and Berendrecht in Stromingen (2015).
[1]:
import os
import pandas as pd
import metran
metran.show_versions()
Python version: 3.10.12 | packaged by conda-forge | (main, Jun 23 2023, 22:40:32) [GCC 12.3.0]
numpy version: 1.26.4
scipy version: 1.12.0
pandas version: 2.2.0
matplotlib version: 3.7.2
pastas version: 1.4.0
numba version: 0.59.0
lmfit version: 1.2.2
Read example data
Read residuals from time series analysis models for 5 piezometers at different depths at location B21B0214. (The time series models are not shown here, only the resulting residuals.)
[2]:
residuals = {}
rfiles = [
os.path.join("./data", f) for f in os.listdir("./data") if f.endswith("_res.csv")
]
for fi in rfiles:
name = fi.split(os.sep)[-1].split(".")[0].split("_")[0]
ts = pd.read_csv(
fi, header=0, index_col=0, parse_dates=True, date_format="%Y-%m-%d"
)
residuals[name] = ts
[3]:
# sort names (not necessary, but ensures the order of things)
sorted_names = list(residuals.keys())
sorted_names.sort()
sorted_names
[3]:
['B21B0214001', 'B21B0214002', 'B21B0214003', 'B21B0214004', 'B21B0214005']
Create Metran model
First collect series in a list with their unique IDs.
[4]:
series = []
for name in sorted_names:
ts = residuals[name]
ts.columns = [name]
series.append(ts)
Create the Metran model and solve.
[5]:
mt = metran.Metran(series, name="B21B0214")
mt.solve()
INFO: Number of factors according to Velicer's MAP test: 1
Fit report B21B0214 Fit Statistics
=======================================================
tmin None obj 2332.33
tmax None nfev 77
freq D AIC 2344.33
solver ScipySolve
Parameters (6 were optimized)
=======================================================
optimal stderr initial vary
B21B0214001_sdf_alpha 5.501017 ±18.98% 10.0 True
B21B0214002_sdf_alpha 13.560042 ±10.04% 10.0 True
B21B0214003_sdf_alpha 4.682870 ±28.86% 10.0 True
B21B0214004_sdf_alpha 11.381674 ±18.22% 10.0 True
B21B0214005_sdf_alpha 13.140605 ±8.48% 10.0 True
cdf1_alpha 22.980925 ±7.43% 10.0 True
Parameter correlations |rho| > 0.5
=======================================================
None
Metran report B21B0214 Factor Analysis
=============================================
tmin None nfct 1
tmax None fep 88.32%
freq D
Communality
=============================================
B21B0214001 73.61%
B21B0214002 87.59%
B21B0214003 93.35%
B21B0214004 91.74%
B21B0214005 81.15%
State parameters
=============================================
phi q
B21B0214001_sdf 0.833781 0.080429
B21B0214002_sdf 0.928908 0.017023
B21B0214003_sdf 0.807716 0.023102
B21B0214004_sdf 0.915889 0.013316
B21B0214005_sdf 0.926724 0.026607
cdf1 0.957419 0.083349
Observation parameters
=============================================
gamma1 scale mean
B21B0214001 0.857982 5.920523 -0.001924
B21B0214002 0.935874 5.565866 -0.055813
B21B0214003 0.966197 5.702295 -0.001265
B21B0214004 0.957794 5.833851 -0.033373
B21B0214005 0.900857 6.234234 -0.022840
State correlations |rho| > 0.5
=============================================
None
Visualizing and accessing Metran results
The results of the Metran can be visualized using the Metran.plots class.
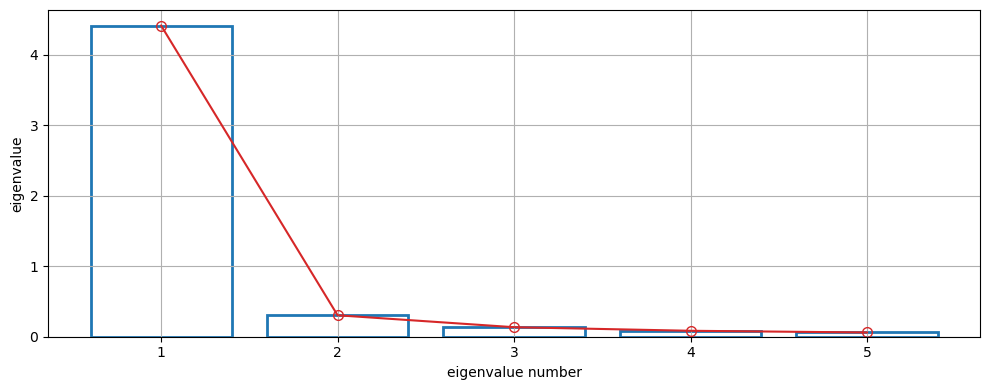
Scree plot
We can draw a scree plot to visualize the eigenvalues (used in determining the number of factors).
[6]:
# Plot eigenvalues in scree plot, see e.g. Fig 2 in JoH paper
ax = mt.plots.scree_plot()
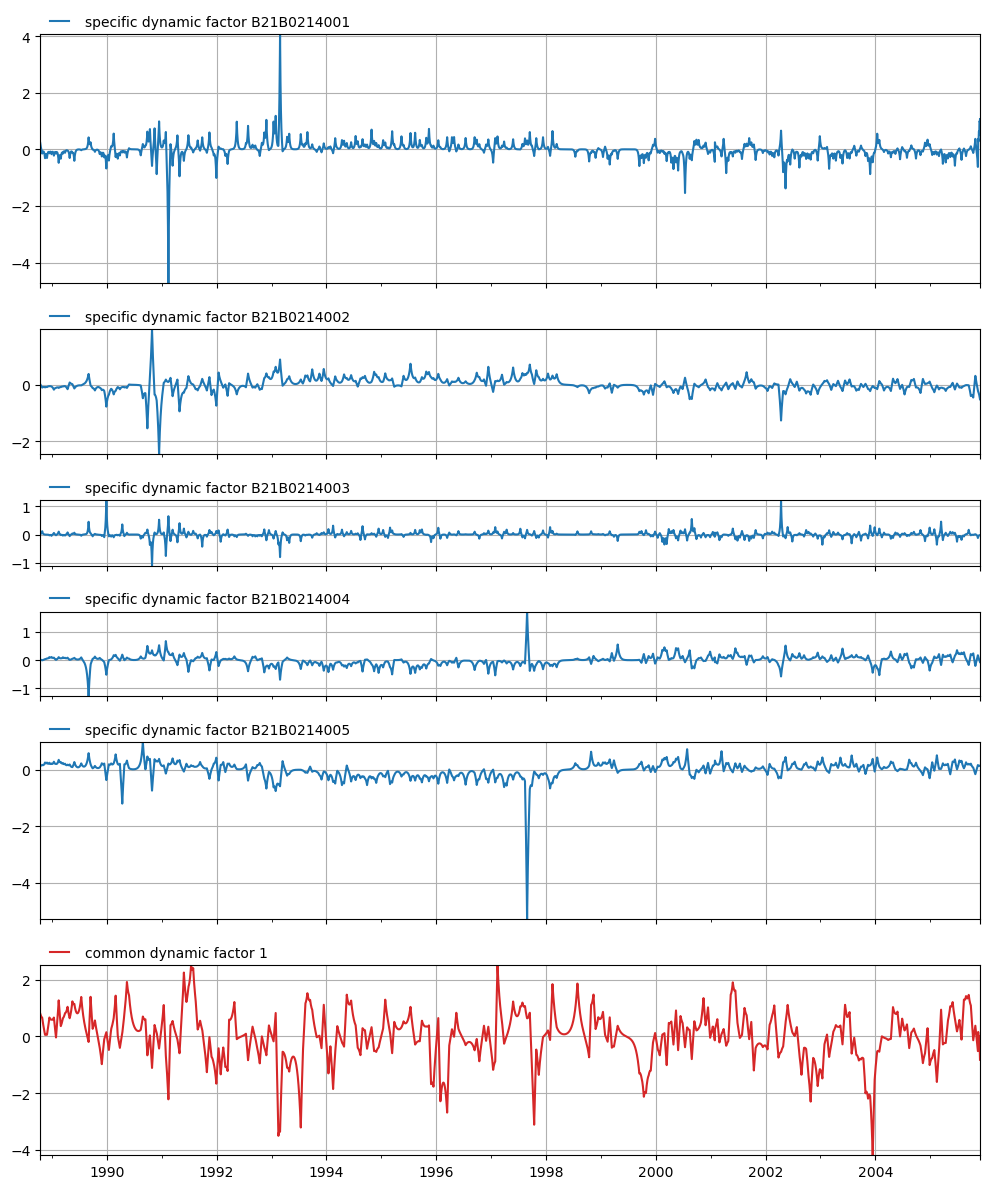
State means
Plot the calculated state means for each of the specific and common dynamic components:
[7]:
mt.get_state_means()
[7]:
| B21B0214001_sdf | B21B0214002_sdf | B21B0214003_sdf | B21B0214004_sdf | B21B0214005_sdf | cdf1 | |
|---|---|---|---|---|---|---|
| date | ||||||
| 1988-10-14 | 0.226549 | 0.021665 | 0.028548 | 0.026005 | 0.153683 | 0.809228 |
| 1988-10-15 | 0.182900 | 0.013039 | 0.026154 | 0.022935 | 0.149910 | 0.790042 |
| 1988-10-16 | 0.145313 | 0.004483 | 0.024958 | 0.020043 | 0.147006 | 0.772353 |
| 1988-10-17 | 0.112540 | -0.004048 | 0.024904 | 0.017306 | 0.144954 | 0.756126 |
| 1988-10-18 | 0.083497 | -0.012601 | 0.025990 | 0.014702 | 0.143742 | 0.741331 |
| ... | ... | ... | ... | ... | ... | ... |
| 2005-11-24 | 0.909463 | -0.389068 | -0.022535 | -0.002155 | 0.133346 | -0.152090 |
| 2005-11-25 | 0.940340 | -0.416086 | -0.021679 | -0.017868 | 0.134755 | -0.288478 |
| 2005-11-26 | 1.001189 | -0.445368 | -0.021816 | -0.033719 | 0.136945 | -0.426332 |
| 2005-11-27 | 0.951887 | -0.477072 | -0.022951 | -0.049831 | 0.139928 | -0.675974 |
| 2005-11-28 | 1.068061 | -0.511373 | -0.025137 | -0.066328 | 0.143722 | -0.823190 |
6255 rows × 6 columns
[8]:
axes = mt.plots.state_means(adjust_height=True)
Simulations
The simulated mean values for each time series in our Metran model can be obtained with:
[9]:
# Get all (smoothed) simulated state means
means = mt.get_simulated_means()
means.head(10)
[9]:
| B21B0214001 | B21B0214002 | B21B0214003 | B21B0214004 | B21B0214005 | |
|---|---|---|---|---|---|
| date | |||||
| 1988-10-14 | 5.450000 | 4.280000 | 4.620000 | 4.640000 | 5.480000 |
| 1988-10-15 | 5.094121 | 4.132049 | 4.500647 | 4.514891 | 5.348733 |
| 1988-10-16 | 4.781724 | 3.992287 | 4.396365 | 4.399175 | 5.231284 |
| 1988-10-17 | 4.505266 | 3.860280 | 4.306655 | 4.292536 | 5.127359 |
| 1988-10-18 | 4.258161 | 3.735609 | 4.231336 | 4.194678 | 5.036712 |
| 1988-10-19 | 4.034568 | 3.617869 | 4.170535 | 4.105327 | 4.959142 |
| 1988-10-20 | 3.829204 | 3.506668 | 4.124708 | 4.024229 | 4.894493 |
| 1988-10-21 | 3.637170 | 3.401621 | 4.094661 | 3.951149 | 4.842653 |
| 1988-10-22 | 3.453793 | 3.302355 | 4.081594 | 3.885870 | 4.803556 |
| 1988-10-23 | 3.274477 | 3.208502 | 4.087163 | 3.828191 | 4.777178 |
For obtaining the data for a simulation with the observations and an (optional) confidence interval, use mt.get_simulation().
[10]:
# Get simulated mean for specific series with/without confidence interval
name = "B21B0214005"
sim = mt.get_simulation(name, alpha=0.05)
sim.head(10)
[10]:
| mean | lower | upper | |
|---|---|---|---|
| date | |||
| 1988-10-14 | 5.480000 | 5.480000 | 5.480000 |
| 1988-10-15 | 5.348733 | 1.669956 | 9.027510 |
| 1988-10-16 | 5.231284 | 0.273555 | 10.189012 |
| 1988-10-17 | 5.127359 | -0.640237 | 10.894955 |
| 1988-10-18 | 5.036712 | -1.263478 | 11.336903 |
| 1988-10-19 | 4.959142 | -1.669234 | 11.587519 |
| 1988-10-20 | 4.894493 | -1.891043 | 11.680029 |
| 1988-10-21 | 4.842653 | -1.942746 | 11.628053 |
| 1988-10-22 | 4.803556 | -1.824438 | 11.431550 |
| 1988-10-23 | 4.777178 | -1.522468 | 11.076824 |
There is also a method to visualize these results for a single time series including optional confidence interval:
[11]:
ax = mt.plots.simulation("B21B0214005", alpha=0.05)
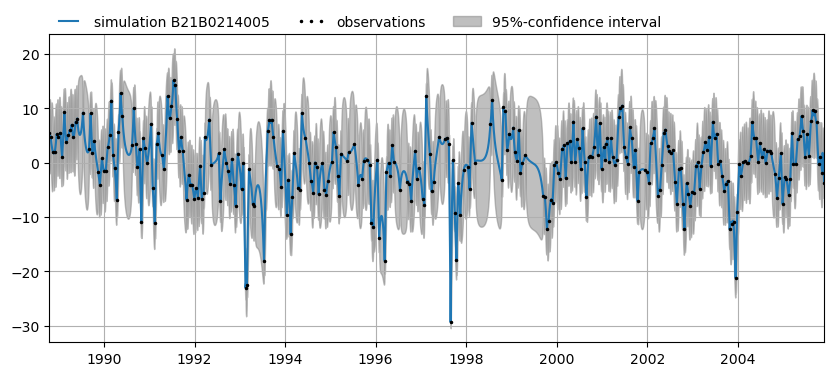
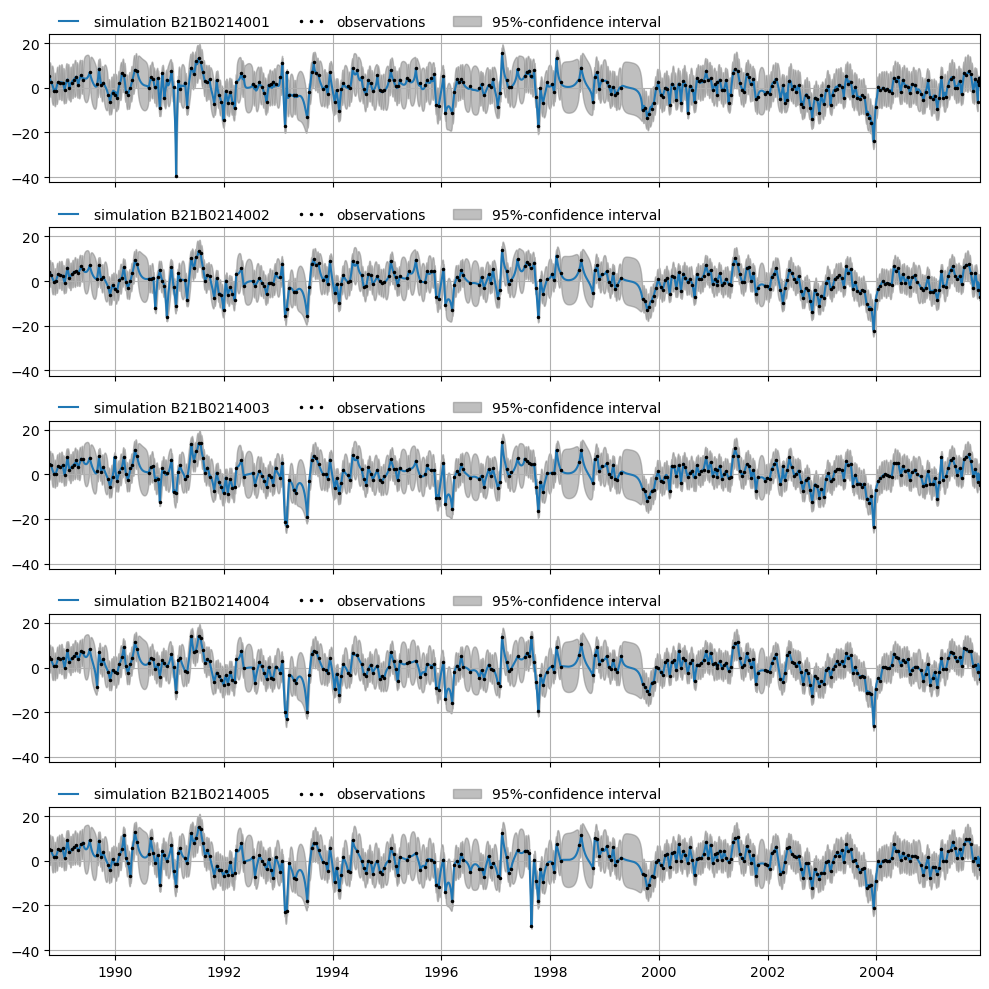
Or all for all time series:
[12]:
axes = mt.plots.simulations(alpha=0.05)
Decompositions
The decomposition of a simulation into specific and common dynamic components can be obtained with mt.decompose_simulation().
[13]:
# Decomposed simulated mean for specific series
decomposition = mt.decompose_simulation("B21B0214001")
decomposition.head(10)
[13]:
| sdf | cdf1 | |
|---|---|---|
| date | ||
| 1988-10-14 | 1.339364 | 4.110636 |
| 1988-10-15 | 1.080942 | 4.013179 |
| 1988-10-16 | 0.858403 | 3.923322 |
| 1988-10-17 | 0.664372 | 3.840894 |
| 1988-10-18 | 0.492420 | 3.765741 |
| 1988-10-19 | 0.336849 | 3.697720 |
| 1988-10-20 | 0.192504 | 3.636701 |
| 1988-10-21 | 0.054601 | 3.582569 |
| 1988-10-22 | -0.081428 | 3.535222 |
| 1988-10-23 | -0.220092 | 3.494570 |
This can also be visualized:
[14]:
axes = mt.plots.decomposition("B21B0214001", split=True, adjust_height=True)
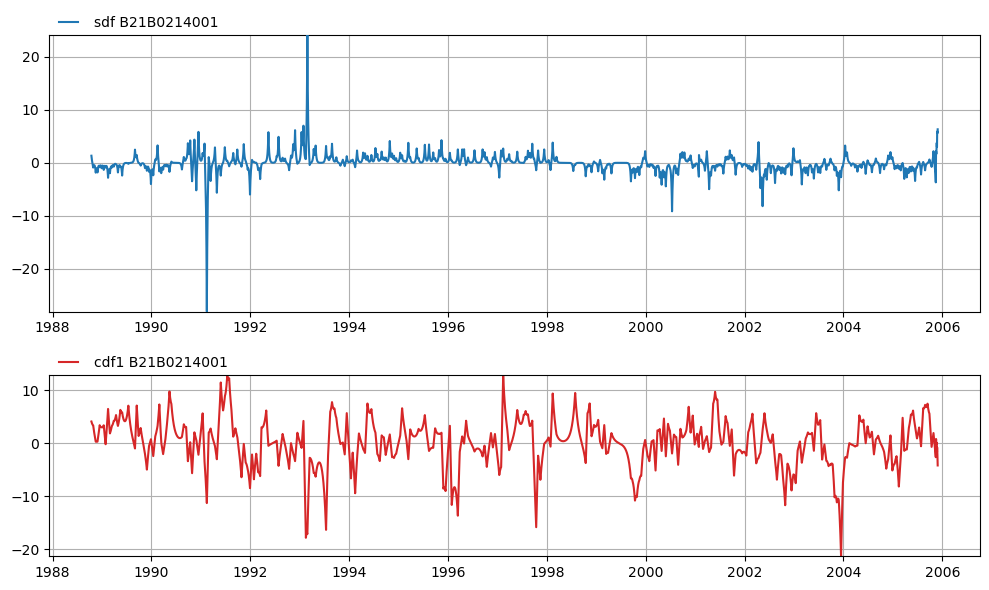
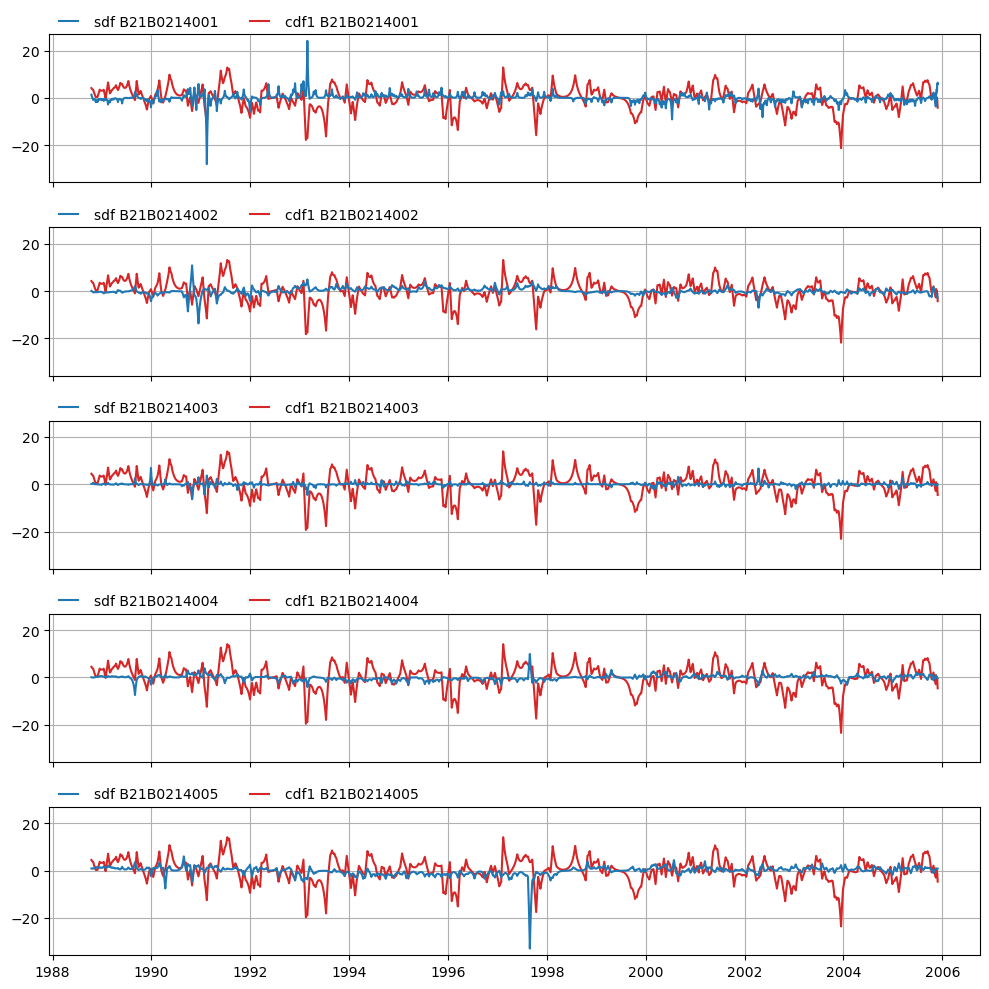
Or for all time series
[15]:
axes = mt.plots.decompositions()
Example application: removing outliers
The Kalman smoother can be re-run after removing (masking) outliers from the observations. This is illustrated below.
First plot the simulation for the original data:
[16]:
name = "B21B0214005"
alpha = 0.05
ax1 = mt.plots.simulation(name, alpha=alpha, tmin="1997-01-01", tmax="1997-12-31")
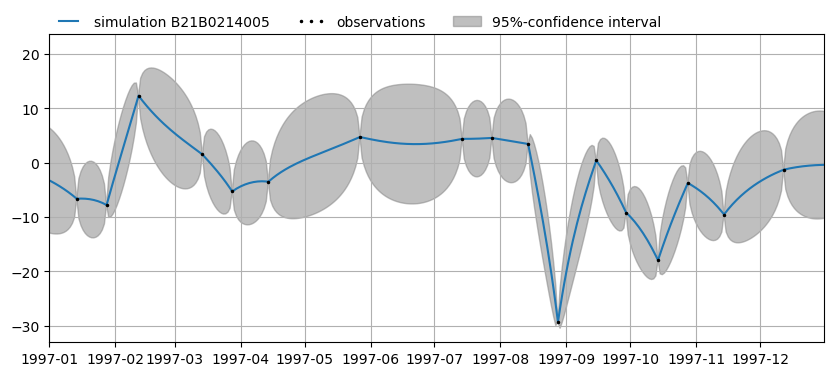
Mask (remove) the outlier on 28 august 1997.
[17]:
oseries = mt.get_observations()
mask = (0 * oseries).astype(bool)
mask.loc["1997-8-28", name] = True
mt.mask_observations(mask)
Now plot the simulation again. Note the estimated value and its 95%-confidence interval for the observation on 28 August 1997 based on the common dynamic factor.
[18]:
# remove outlier from series B21B0214005 at 1997-8-28
# and re-run smoother to get estimate of observation
# (Fig 3 in Stromingen without deterministic component)
ax2 = mt.plots.simulation(name, alpha=alpha, tmin="1997-01-01", tmax="1997-12-31")
sim = mt.get_simulation(name, alpha=None).loc[["1997-8-28"]]
# plot outlier and corrected value
outlier = oseries.loc[["1997-8-28"], name]
ax2.plot(outlier.index, outlier, "C3x", label="outlier")
ax2.plot(sim.index, sim, "C2s", label="corrected value", mec="k", mew=0.5)
ax2.legend(loc=(0, 1), numpoints=1, frameon=False, ncol=3)
INFO: Running Kalman filter with masked observations.
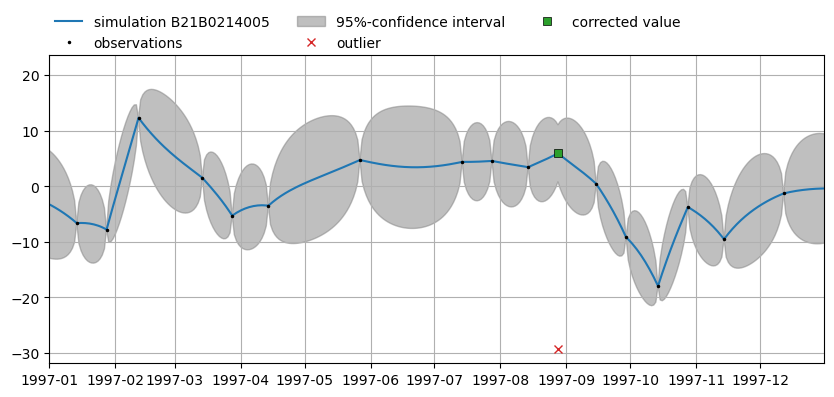
To reset the observations (remove all masks):
[19]:
# unmask observations to get original observations
mt.unmask_observations()
References
Van Geer, F.C. en W.L. Berendrecht (2015) Meervoudige tijdreeksmodellen en de samenhang in stijghoogtereeksen. Stromingen 23 nummer 3, pp. 25-36.