Pastas and Metran example
This notebook shows how output from Pastas time series models can be analyzed using Metran.
[1]:
import os
import hydropandas as hpd
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pastas as ps
import metran
ps.logger.setLevel("ERROR")
metran.show_versions()
Python version: 3.10.12 | packaged by conda-forge | (main, Jun 23 2023, 22:40:32) [GCC 12.3.0]
numpy version: 1.26.4
scipy version: 1.12.0
pandas version: 2.0.3
matplotlib version: 3.8.3
pastas version: 1.4.0
numba version: 0.59.0
lmfit version: 1.2.2
Read data
Load the observed heads from piezometers at different depths at location B21B0214. The outliers (values outside of \(5 \sigma\) (std. dev.)) are removed from the time series.
[2]:
oc = hpd.read_dino("./data", subdir=".")
[3]:
oc
[3]:
| x | y | filename | source | unit | monitoring_well | tube_nr | screen_top | screen_bottom | ground_level | tube_top | metadata_available | obs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| name | |||||||||||||
| B21B0214-001 | 198085.0 | 518413.0 | B21B0214001_1 | dino | m NAP | B21B0214 | 1.0 | -7.3 | -9.3 | -0.29 | 0.24 | True | GroundwaterObs B21B0214-001 -----metadata-----... |
| B21B0214-003 | 198085.0 | 518413.0 | B21B0214003_1 | dino | m NAP | B21B0214 | 3.0 | -24.3 | -26.3 | -0.29 | 0.25 | True | GroundwaterObs B21B0214-003 -----metadata-----... |
| B21B0214-002 | 198085.0 | 518413.0 | B21B0214002_1 | dino | m NAP | B21B0214 | 2.0 | -15.3 | -17.3 | -0.29 | 0.28 | True | GroundwaterObs B21B0214-002 -----metadata-----... |
| B21B0214-004 | 198085.0 | 518413.0 | B21B0214004_1 | dino | m NAP | B21B0214 | 4.0 | -36.3 | -38.3 | -0.29 | 0.23 | True | GroundwaterObs B21B0214-004 -----metadata-----... |
| B21B0214-005 | 198085.0 | 518413.0 | B21B0214005_1 | dino | m NAP | B21B0214 | 5.0 | -51.3 | -53.3 | -0.29 | 0.21 | True | GroundwaterObs B21B0214-005 -----metadata-----... |
[4]:
oseries = {}
for o in oc.obs:
name = o.name
o = o["stand_m_tov_nap"].rename(o.name)
# remove outliers outside 5*std
mean = o.median()
std = o.std()
mask_outliers = (o - mean).abs() > 5 * std
ts = o.copy()
ts.loc[mask_outliers] = np.nan
# store time series
oseries[name] = ts
[5]:
# sort the names
sorted_names = list(oseries.keys())
sorted_names.sort()
sorted_names
[5]:
['B21B0214-001',
'B21B0214-002',
'B21B0214-003',
'B21B0214-004',
'B21B0214-005']
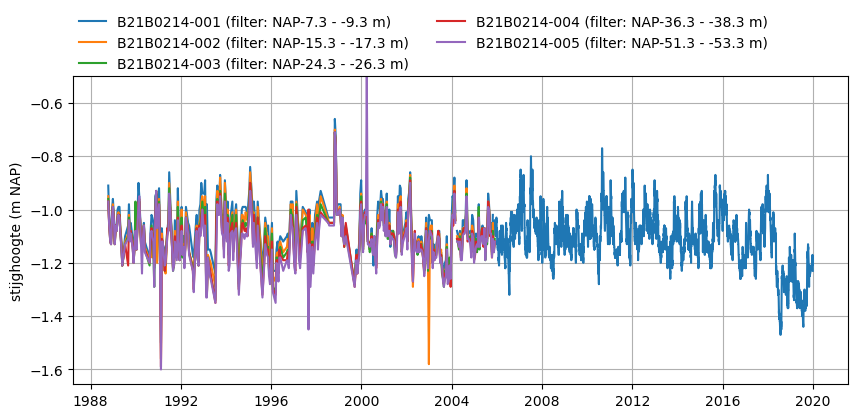
Plot the heads:
[6]:
fig, ax = plt.subplots(1, 1, figsize=(10, 4))
for name in sorted_names:
o = oseries[name]
ftop = oc.loc[name, "screen_top"]
fbot = oc.loc[name, "screen_bottom"]
lbl = f"{name} (filter: NAP{ftop:+.1f} - {fbot:+.1f} m)"
ax.plot(o.index, o, label=lbl)
ax.set_ylabel("stijghoogte (m NAP)")
ax.legend(loc=(0, 1), ncol=2, frameon=False)
ax.set_ylim(top=-0.5)
ax.grid(True)
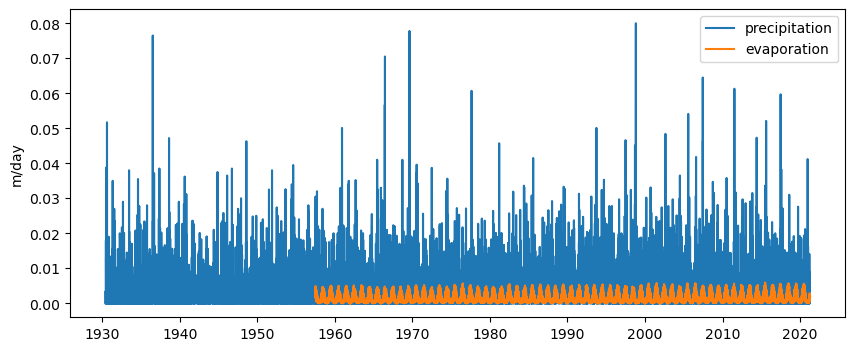
Load the precipitation and evaporation data from two nearby weather stations
[7]:
p = pd.read_csv(
"./data/RD_338.csv", index_col=[0], parse_dates=True, usecols=["YYYYMMDD", "RD"]
)
e = pd.read_csv(
"./data/EV24_260.csv", index_col=[0], parse_dates=True, usecols=["YYYYMMDD", "EV24"]
)
Plot precipitation and evaporation time series
[8]:
fig, ax = plt.subplots(1, 1, figsize=(10, 4))
ax.plot(p.index, p, label="precipitation")
ax.plot(e.index, e, label="evaporation")
ax.set_ylabel("m/day")
ax.legend(loc="best")
[8]:
<matplotlib.legend.Legend at 0x7fe032f56770>
Build time series models
The time series models attempt to simulate the heads using recharge as stress. The recharge is calculated using \(R = P - f \cdot E\), where \(R\) is recharge, \(P\) is precipitation, \(E\) is evaporation and \(f\) is factor that is optimized. The model fit results are printed to the console. The model residuals are stored for analysis with Metran.
[9]:
# Normalize the index (reset observation time to midnight (the end of the day)).
p.index = p.index.normalize()
e.index = e.index.normalize()
# set tmin/tmax
tmin = "1988-10-14"
tmax = "2005-11-28"
# store models and residuals
models = []
residuals = []
for name in sorted_names:
# create model
ml = ps.Model(oseries[name])
rm = ps.RechargeModel(prec=p, evap=e)
ml.add_stressmodel(rm)
# solve model
ml.solve(tmin=tmin, tmax=tmax, report=False)
# print fit statistic
print(name, f"EVP = {ml.stats.evp():.1f}%")
# store model
models.append(ml)
# get residuals
r = ml.residuals()
r.name = name
residuals.append(r)
B21B0214-001 EVP = 68.4%
B21B0214-002 EVP = 58.9%
B21B0214-003 EVP = 66.8%
B21B0214-004 EVP = 61.3%
B21B0214-005 EVP = 51.0%
Build Metran model
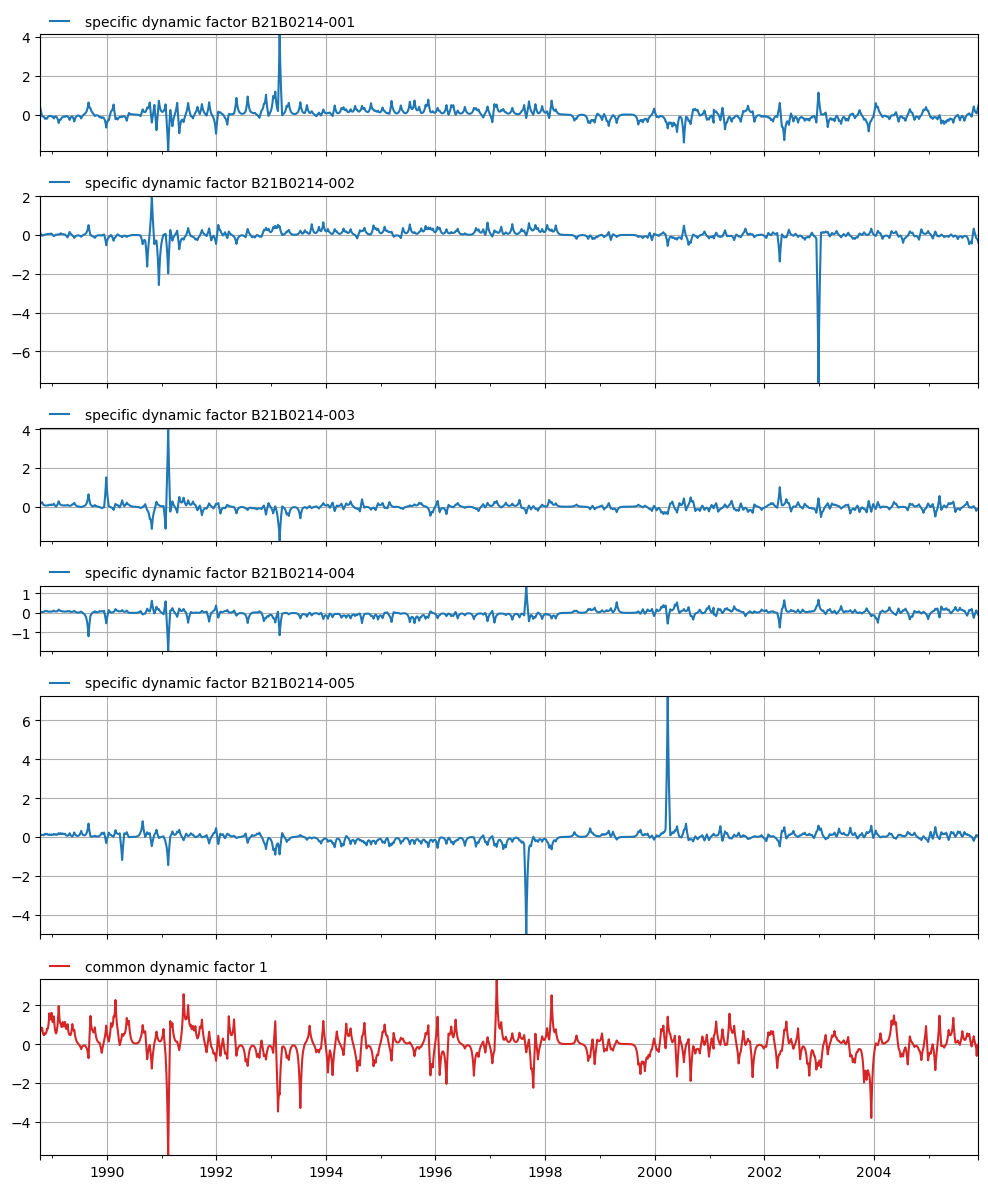
A Metran model is created using the residuals of the time series models. By analyzing the model residuals we can determine for example, whether there is a common pattern in the residuals, which could indicate a missing influence, or a shortcoming in the model structure. Additionally we might be able to analyze whether there are still outliers left in our time series.
[10]:
mt = metran.Metran(residuals)
mt.solve()
INFO: Number of factors according to Velicer's MAP test: 1
Fit report Cluster Fit Statistics
======================================================
tmin None obj 3223.12
tmax None nfev 7
freq D AIC 3235.12
solver ScipySolve
Parameters (6 were optimized)
======================================================
optimal stderr initial vary
B21B0214-001_sdf_alpha 10 ±10.00% 10 True
B21B0214-002_sdf_alpha 10 ±10.00% 10 True
B21B0214-003_sdf_alpha 10 ±10.00% 10 True
B21B0214-004_sdf_alpha 10 ±10.00% 10 True
B21B0214-005_sdf_alpha 10 ±10.00% 10 True
cdf1_alpha 10 ±10.00% 10 True
Parameter correlations |rho| > 0.5
======================================================
None
Metran report Cluster Factor Analysis
==============================================
tmin None nfct 1
tmax None fep 81.32%
freq D
Communality
==============================================
B21B0214-001 84.36%
B21B0214-002 67.77%
B21B0214-003 88.55%
B21B0214-004 90.71%
B21B0214-005 63.38%
State parameters
==============================================
phi q
B21B0214-001_sdf 0.904837 0.028355
B21B0214-002_sdf 0.904837 0.058418
B21B0214-003_sdf 0.904837 0.020764
B21B0214-004_sdf 0.904837 0.016846
B21B0214-005_sdf 0.904837 0.066383
cdf1 0.904837 0.181269
Observation parameters
==============================================
gamma1 scale mean
B21B0214-001 0.918464 0.052053 -0.000691
B21B0214-002 0.823243 0.060746 -0.000529
B21B0214-003 0.940984 0.050673 -0.000204
B21B0214-004 0.952399 0.056584 0.000345
B21B0214-005 0.796109 0.070582 0.000577
State correlations |rho| > 0.5
==============================================
None
Plot the specific and common dynamic components
[11]:
axes = mt.plots.state_means()
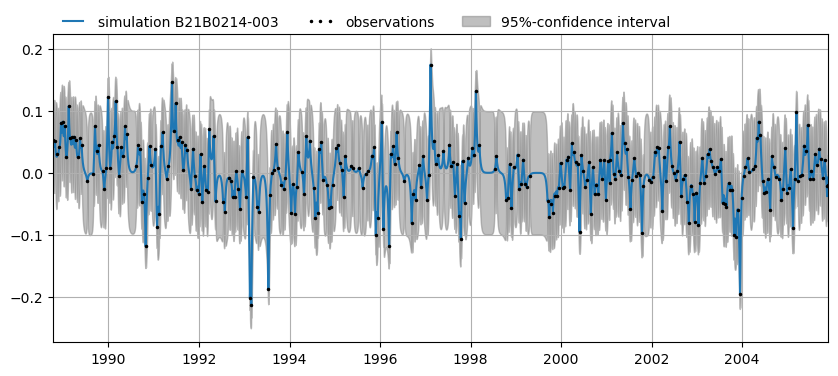
Plot a simulation, including a confidence interval for B21B0214003.
[12]:
ax = mt.plots.simulation(mt.snames[2])